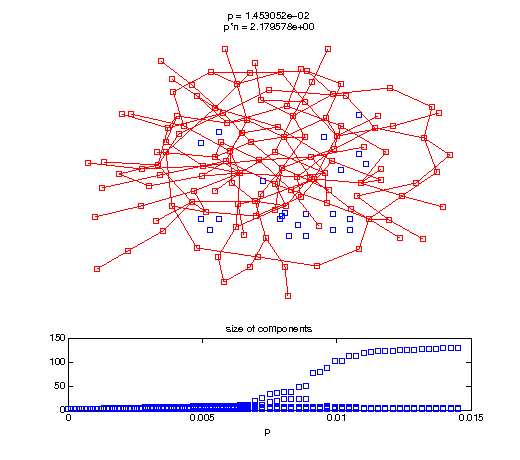
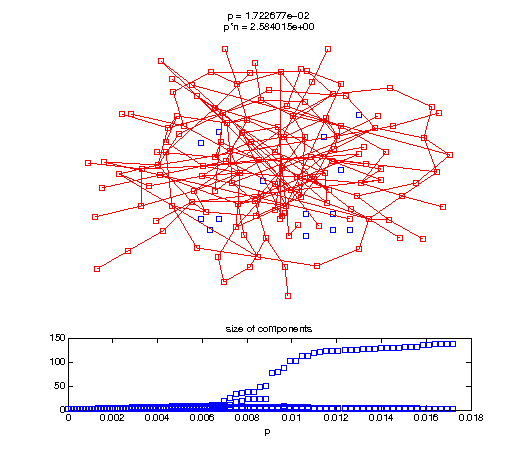
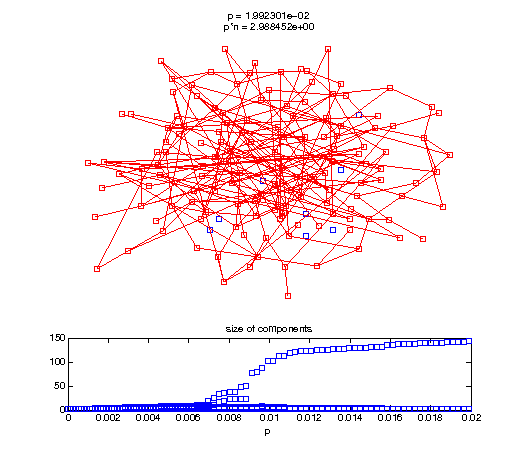
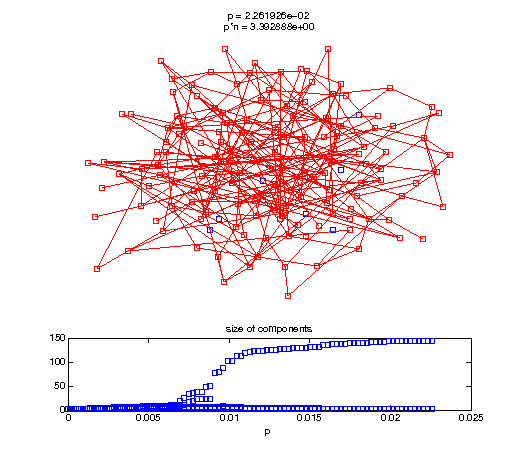
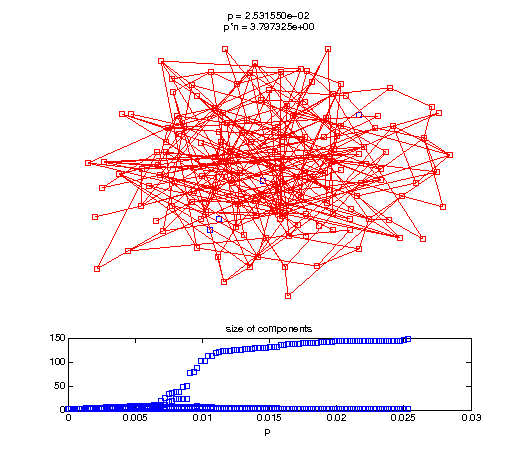
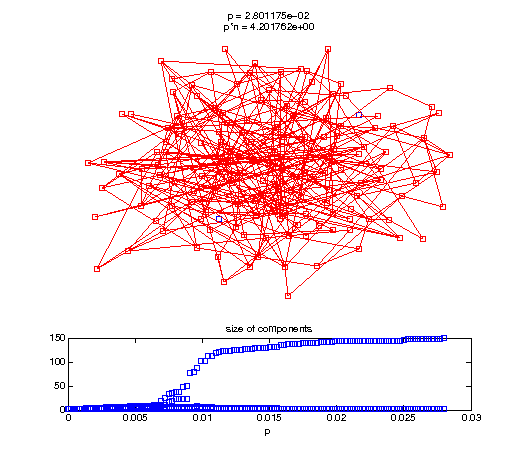
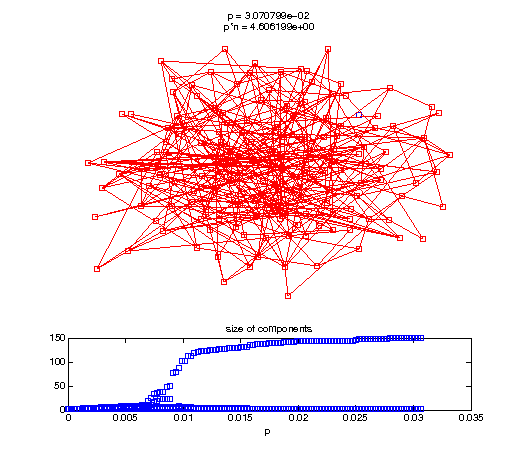
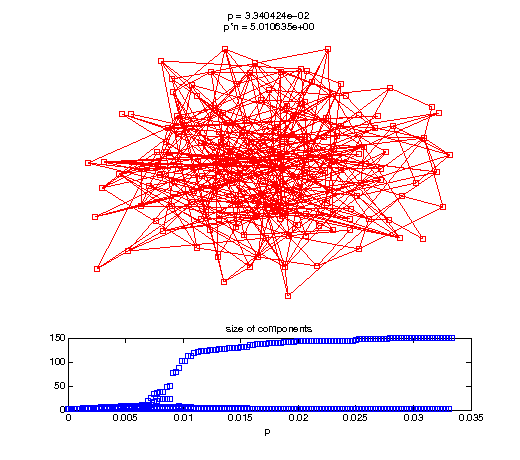
Evolution of the giant connected component
In the last example, we saw how an Erdos-Renyi graph has a giant connected component when the connection probability exceeds a threshold. In this script, we will watch the evolution process closely. This script is designed to be run locally, because it produces an animated figure with many subplots. The HTML fill will produce a few presentative examples.
Contents
Parameterize the Erdos-Renyi graph
Here, we first create a parametric representation of varying the connection probability in an Erdos-Renyi random graph.
rand('seed',100); n = 150; A = rand(n); A = triu(A,1); A = A + A'; % make sure the seed data is symmetric % define a function to produce a graph given a probability G = @(p) A < p;
Define thresholds for the Erdos-Renyi graph
% define the thresholds where the graph is fully connected and has % a giant component thres_conn = log(n)/(n); thres_giant = 1/(n-1); % define a layout threshold at the geometric mean of these thresholds thres = sqrt(thres_conn*thres_giant);
Compute a static layout
find the layout at the given threshold
Gt = G(thres); [x y] = neato_layout(Gt);
Show the graph's evolution
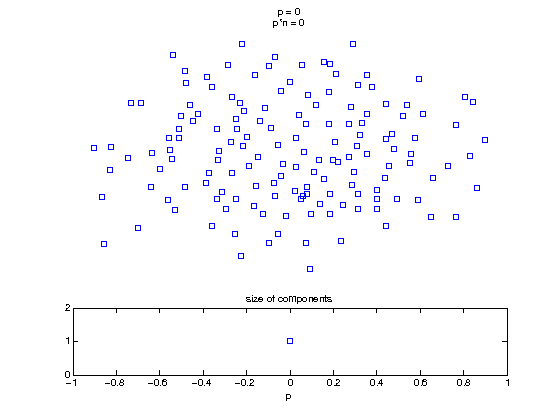
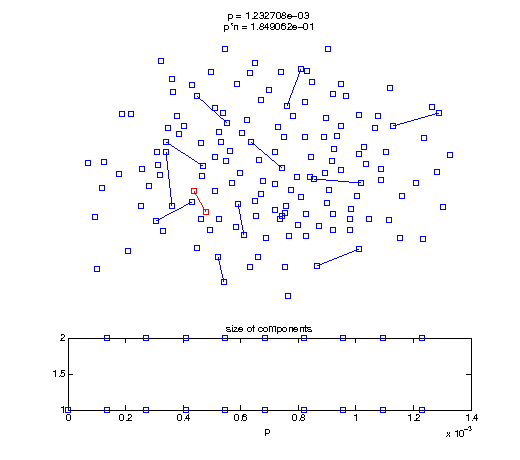
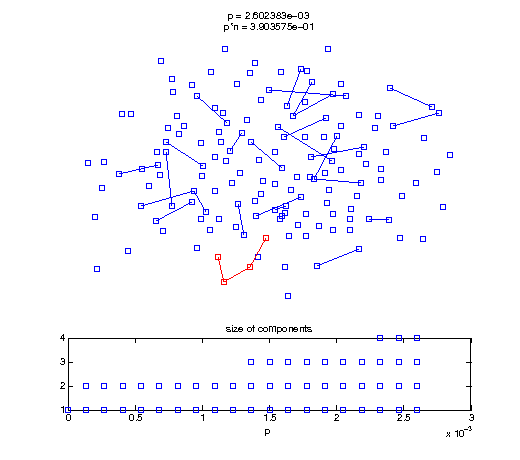
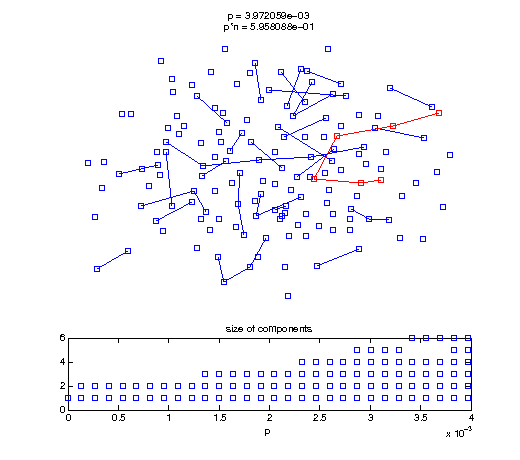
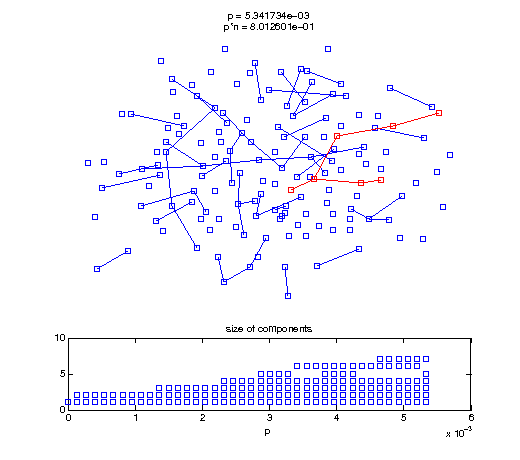
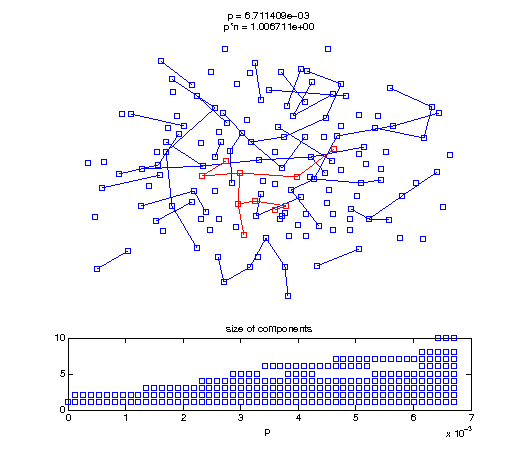
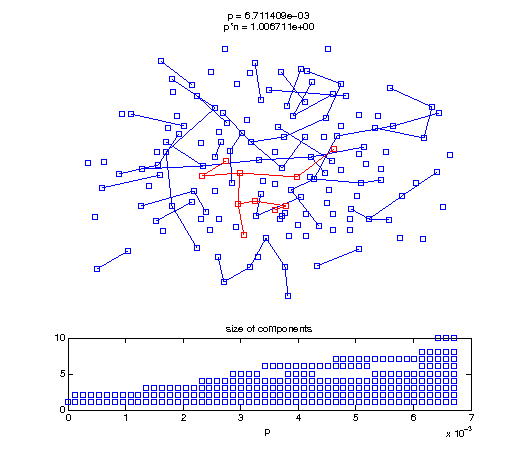
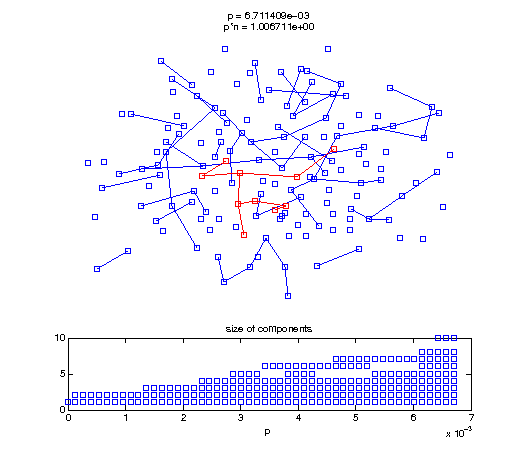
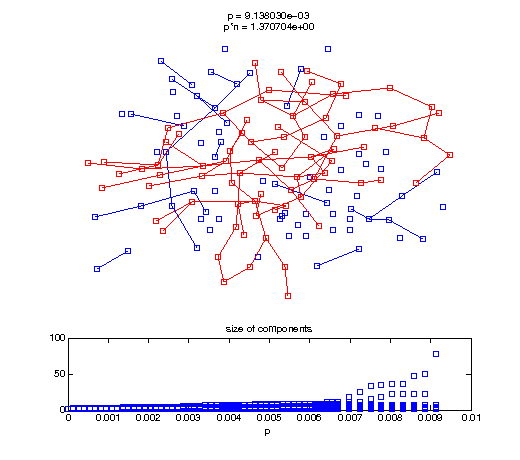
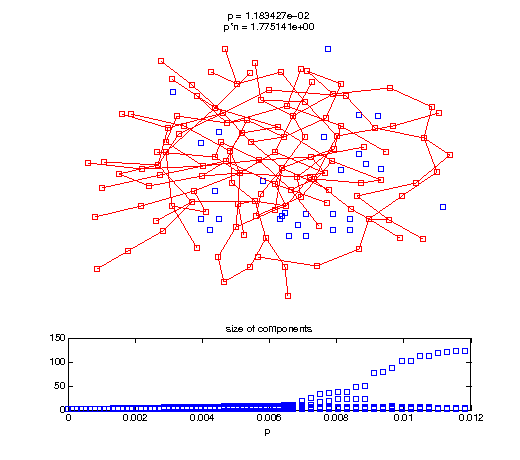
Now, we'll watch as the graph evolves as we keep adding edges by increasing the probability p through all the various thresholds.
Define a sequence of probabilities to explore.
t1 = linspace(0, thres_giant, 50); t2 = linspace(thres_giant, thres_conn, 100); % we want to pause things at the emergence of the giant component steps = [t1 ones(1,40)*thres_giant t2]; nframes = length(steps); comp_size_history = []; comp_step_history = []; for frame=1:nframes p = steps(frame); Gcur = G(p); % make one large subplot for the graph layout with the % giant component highlighted subplot(4,2,[1 2 3 4 5 6]); gplot(Gcur, [x' y'],'bo-'); hold on; plot(x', y', 'bo'); [Glc pcc] = largest_component(Gcur); gplot(Glc, [x(pcc)' y(pcc)'], 'ro-'); hold off; axis off; title({sprintf('p = %d', p), sprintf('p*n = %d', p*n)}); % make a smaller subplot for the size of the components subplot(4,2,[7 8]); [comps comp_sizes] = components(Gcur); % get the sizes comp_size_history = [comp_size_history comp_sizes]; comp_step_history = [comp_step_history ones(1,length(comp_sizes))*p]; plot(comp_step_history, comp_size_history, 'o'); title('size of components'); xlabel('p'); set(gcf,'Color',[1,1,1]); % pause for display pause(0.15); if mod(frame,10)==0 || frame==1 || frame==nframes, snapnow; end end